
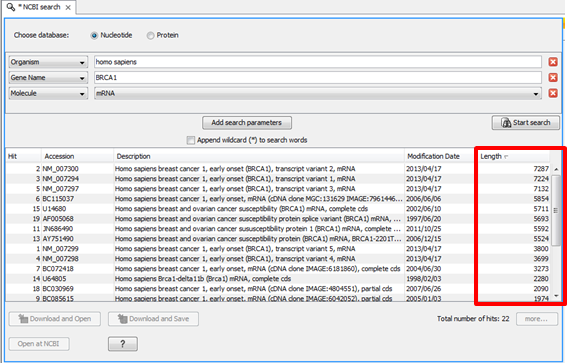
All additional columns are added to the right of the alignment. The dialog will only suggest a column if one or more sequences in the alignment report that metadata. Optional columns may include Gene (gene symbols for RefSeq accessions), Date (collection date), Country (collection country), Host (pathogen host), or Source (tissue source). This option opens a dialog where you can customize the columns shown. These columns can be removed and/or additional columns added using the Columns option on the toolbar. sequence accession), Start and End of the alignment, and Organism (species name). Note: for the image on this page, the strand indicator ((+) or (-)) was hidden by adding the parameter: &columns=s:0 to the URL.īy default, the Alignment view shows metadata columns for Sequence ID (e.g. Note: Organism information is not shown for user-provided sequences without an NCBI accession or for NCBI accessions reported with range, e.g. The species associated with the sequence accession is reported in the Organism column. NCBI accession identifiers are linked to the corresponding resource page in the NCBI SRA, Nucleotide or Protein databases. The Sequence ID column shows sequence identifiers from the alignment.
CLC SEQUENCE VIEWER ADD ANNOTATION DOWNLOAD
Please download the 16SRNA_Deino_87seq.aln data file for this tutorial.
CLC SEQUENCE VIEWER ADD ANNOTATION HOW TO
This guide will show you how to upload data into MSA viewer and perform basic operations including navigation, setting an anchor row, hiding rows, and changing the coloration method. The MSA home page includes a links to sample protein and DNA alignment sessions. Users can also upload and view their own alignment files in alignment FASTA or ASN format. Multiple Sequence Alignment Viewer application (MSA) is a web application that visualizes alignments created by programs such as MUSCLE or CLUSTAL, including alignments from NCBI BLAST results.

Guide to Using the Multiple Sequence Alignment Viewer


 0 kommentar(er)
0 kommentar(er)
